Peptide Repertoire-Based Anchor Motif
An application to define HLA binding motifs
How to use PRBAM v1.0
- The user must select the MHC-I or HLA-DR module.
- Select a peptide list (text or fasta file). The peptide sequences must be in one-letter amino acid code. Only the 20 regular amino acids are considered for the final score.
- Click on the “Submit” button.
Allowed file formats
Plain text file (example.txt)
[...]
QVAEYKQLKDTLNRIPS
VAEYKQLKDTLNRIPS
EEFLSFLHPEHSRG
KGRLDYLSSLKVKGL
IRPDGEKKAYVRLAPDYDALDVA
EKKAYVRLAPDYDALD
EKKAYVRLAPDYDAL
HHTFYNELRVAPEEHPV
[...]
Fasta file (example.fasta)
[...]
>sequence 12
QVAEYKQLKDTLNRIPS
>sequence 13
VAEYKQLKDTLNRIPS
>sequence 14
EEFLSFLHPEHSRG
>sequence 15
KGRLDYLSSLKVKGL
>sequence 16
IRPDGEKKAYVRLAPDYDALDVA
>sequence 17
EKKAYVRLAPDYDALD
>sequence 18
EKKAYVRLAPDYDAL
>sequence 19
HHTFYNELRVAPEEHPV
[...]
Example

Select the module (MHC-I or HLA-DR) and select a peptide list (text or fasta file). Then click on Submit button.
Chart tab shows a representation of the DMP values of each amino acid in each position.
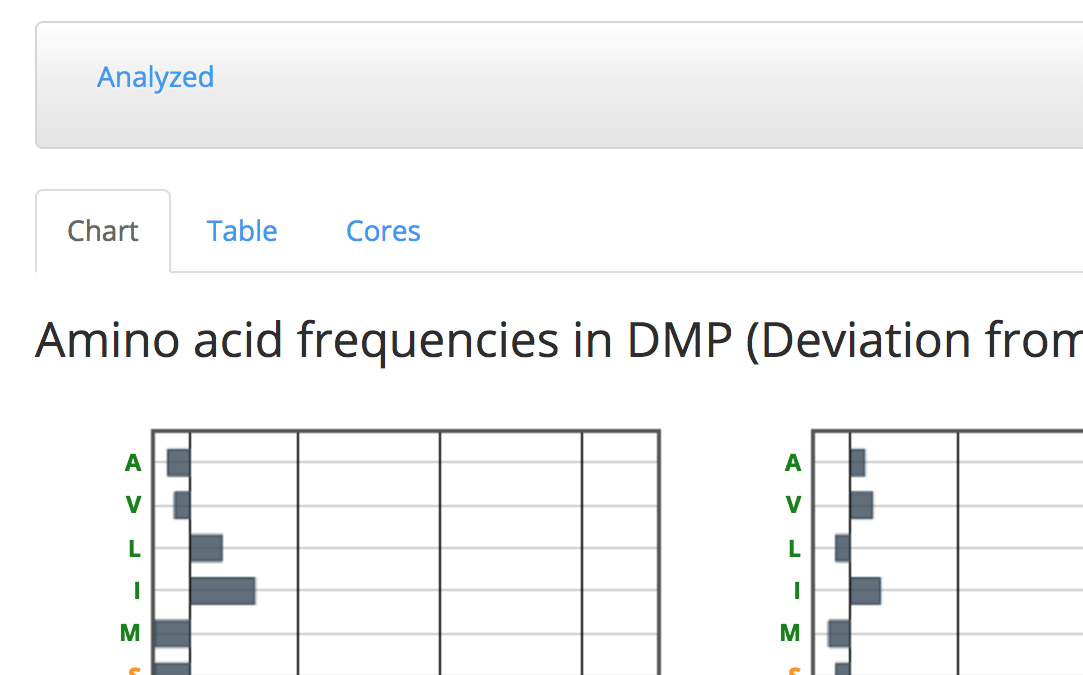
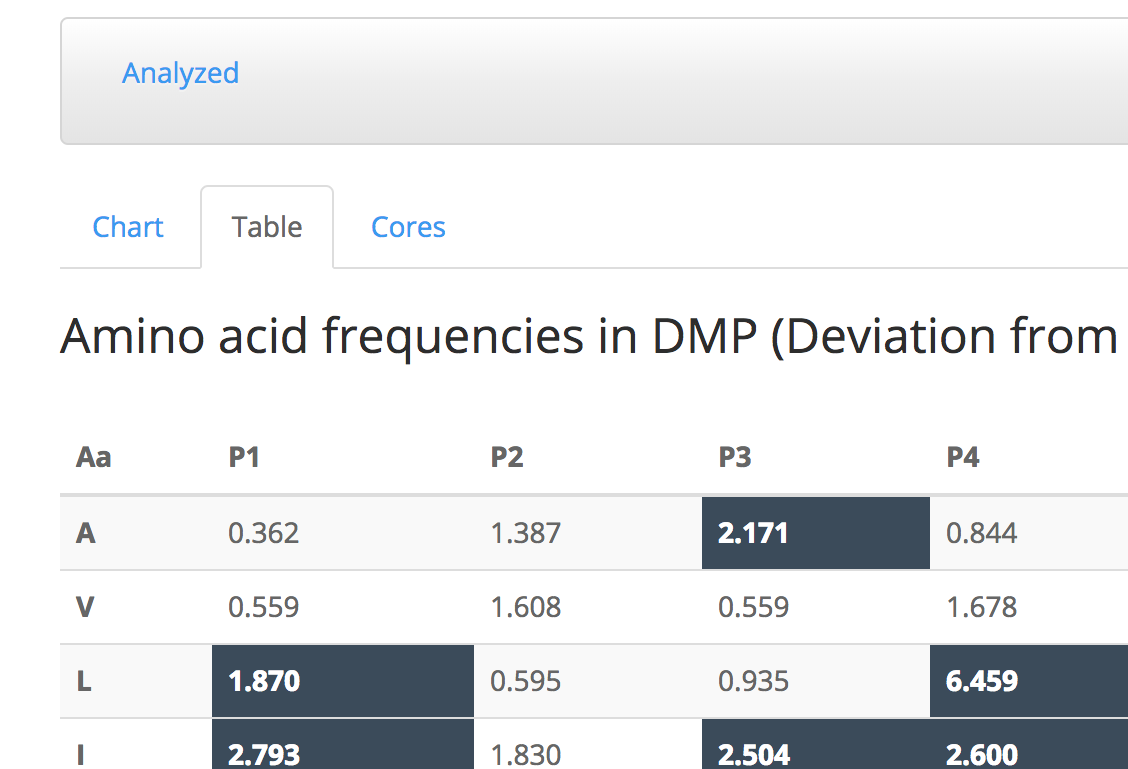
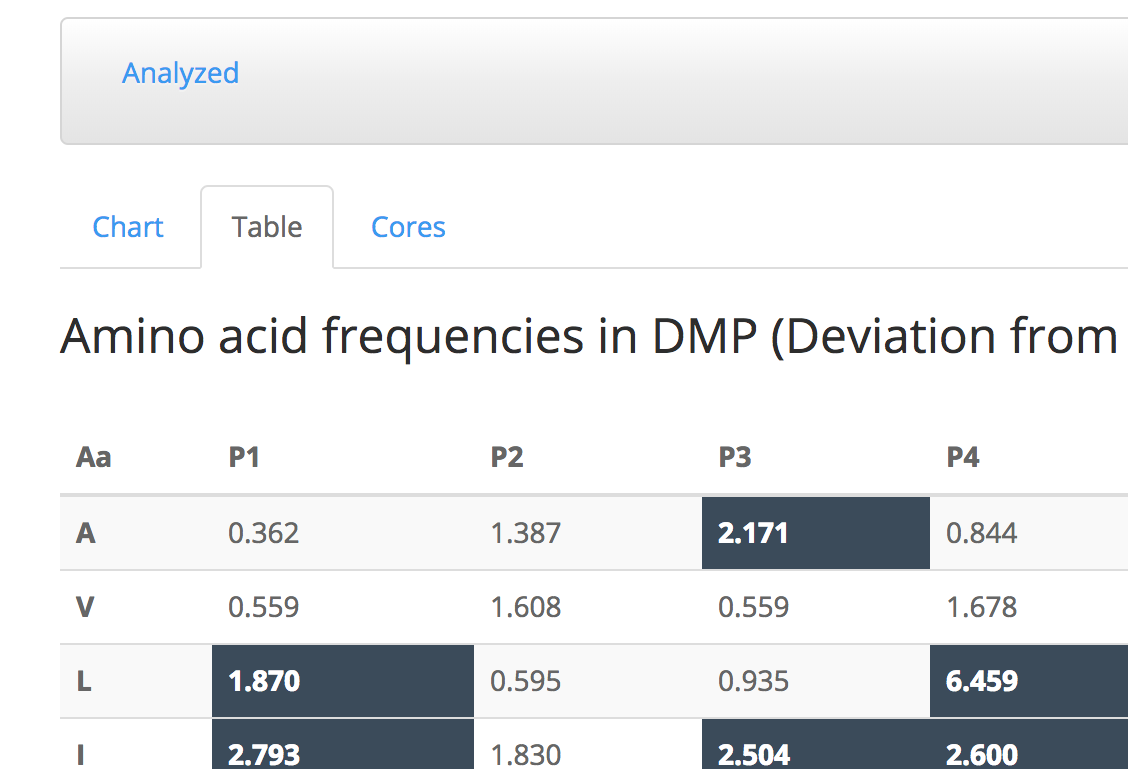
Table tab shows the DMP values of each amino acid in each position in a table format.
Cores tab shows the list of all peptides processed by PRBAM. The selected cores are highlighted and listed with their corresponding scores.